
Mass Spec Query Language Support to the Spectra Package
Source:vignettes/SpectraQL.Rmd
SpectraQL.RmdPackage: SpectraQL
Authors: Johannes Rainer [aut, cre] (ORCID: https://orcid.org/0000-0002-6977-7147), Andrea Vicini
[aut], Sebastian Gibb [ctb] (ORCID: https://orcid.org/0000-0001-7406-4443)
Compiled: Fri May 16 19:49:52 2025
Introduction
The Mass Spec Query Language (MassQL) is a domain specific language meant to be a succinct way to express a query in a mass spectrometry (MS) centric fashion. It is inspired by SQL, but it attempts to bake in assumptions of MS to make querying much more natural for MS users.
The SpectraQL package provides support for the MassQL
language in R, for MS data represented by Spectra objects
defined in Bioconductor’s Spectra
package.
Installation
The package can be installed with the BiocManager
package using:
## Install BiocManager, if not already installed
install.packages("BiocManager")
## Install the package
BiocManager::install("SpectraQL")Extracting data from Spectra objects with MassQL
The SpectraQL package adds support for most of the Mass Spec
Query Language functionality to Spectra objects hence
allowing to filter and extract data using platform independent and data
storage agnostic queries. At present, SpectraQL supports most,
but not all of the conditions and expressions of the MassQL definition.
In particular, numeric operations in query fields such as
MS2PROD=100+14 are not yet supported.
Below we load MS data from an example mzML file. Note also that
Spectra objects can represent MS data from a very large
variety of sources including but not limited to mzML or mzXML files,
databases (e.g. MsBackendMassbank)
or MGF files (MsBackendMgf).
The use of the Spectra
an hence SpectraQL package is thus not restricted to
mzML-backed MS data.
library(Spectra)
library(SpectraQL)
library(msdata)
fl <- system.file("TripleTOF-SWATH", "PestMix1_DDA.mzML", package = "msdata")
dda <- Spectra(fl)MassQL definition
A MassQL query consists of several parts:
"QUERY <type of data> WHERE <condition> AND <condition> FILTER <filter> AND <filter>"
(see its definition
for more details). "<type of data>" defines which
data should be extracted from the selected data set,
"<condition>" defines which spectra should be
selected and "<filter>" allows to subset each
individual spectrum (i.e. to which mass peaks each spectrum should be
subsetted). See the following sections for more information on each part
of such an MassQL query. SpectraQL supports the MassQL query
schema is however case insensitive (i.e. both "query" and
"QUERY" are accepted) and is insensitive to white spaces
(i.e. both "RTMIN=10" and "RTMIN = 10" are
supported). Also, it should be noted that not all keywords, filters and
operations defined by MassQL are yet supported by SpectraQL.
The supported types of data, conditions and filters are listed in the
sections below.
Type of data
The "<type of data>" defines what should
be returned by the query function. In addition, functions
can be specified and applied to "<type of data>" to
summarize the results. The types of data that are currently supported by
SpectraQL are:
-
"*": returns the full data, i.e. returns aSpectraobject. -
"MS1DATA"(case insensitive): returns (if no function is defined; see further below) the peaks data for selected MS1 scans (i.e. alistof matrices with m/z and intensity values). -
"MS2DATA"(case insensitive): returns (if no function is defined) the peaks data for selected MS2 scans (i.e. alistof matrices with m/z and intensity values).
In addition, functions can be used to extract specific information from the selected spectra. SpectraQL supports at present the following functions defined by MassQL:
-
"scaninfo": e.g."scaninfo(MS1DATA)"or"scaninfo(MS2DATA)"to extract spectra information for MS1 or MS2 scans, respectively. This returns thespectraDatafor the sub-settedSpectraobject. -
"scansum": e.g."scansum(MS1DATA)"or"scansum(MS2DATA)"to extract the TIC (sum of peak intensities) of the selected spectra.
Condition
The "<condition>" allows to subset a
Spectra object. Several conditions can be combined with
"and". The syntax for a condition is
"<condition> = <value>",
e.g. "MS2PROD = 144.1". Such conditions can be further
refined by additional expressions that allow for example to define
acceptable tolerances for m/z differences (see further below for
details). SpectraQL supports the following conditions:
-
"RTMIN": minimum retention time (in seconds). -
"RTMAX": maximum retention time (in seconds). -
"SCANMIN": the minimum scan number (acquisition number). -
"SCANMAX": the maximum scan number (acquisition number). -
"CHARGE": the charge for MS2 spectra. -
"POLARITY": the polarity of the spectra (can be"positive","negative","pos"or"neg", case insensitive). -
"MS2PROD"or"MS2MZ": allows to select MS2 spectra that contain a peak with a particular m/z. -
"MS2PREC": allows to select MS2 spectra with the defined precursor m/z. -
"MS1MZ": allows to select MS1 spectra containing peaks with the defined m/z. -
"MS2NL": allows to look for a neutral loss from precursor in MS2 spectra.
All conditions involving m/z values allow to specify a mass accuracy
using the optional fields "TOLERANCEMZ" and
"TOLERANCEPPM" that define the absolute and m/z-relative
acceptable difference in m/z values. One or both fields can be attached
to a condition such as
"MS2PREC=100:TOLERANCEMZ=0.1:TOLERANCEPPM=20" to select for
example all MS2 spectra with a precursor m/z equal to 100 accepting a
difference of 0.1 and 20 ppm. Note that in contrast to MassQL, the
default tolarance and ppm is 0 for all calls.
Filter
Filters allow to subset individual spectra keeping e.g. only peaks that match a user-defined m/z value. SpectraQL supports the following filters:
-
"MS1MZ": filters MS1 spectra keeping only peaks with matching m/z values (tolerance can be specified as above with"TOLERANCEMZ"and"TOLERANCEPPM". -
"MS2MZ": filters MS2 spectra keeping only peaks with matching m/z values (tolerance can be specified as above with"TOLERANCEMZ"and"TOLERANCEPPM".
Examples
In this section we use MassQL queries to subset and extract data from
our Spectra object dda. This object contains
MS1 and MS2 spectra from a data dependent acquisition. Some general
information is provided below.
##
## 1 2
## 4627 2975## [1] 0.231 899.993Filtering and subsetting
To restrict the data to spectra measured between 200 and 300 seconds
of the measurement run we can use a MassQL query with the
"rtmin" and "rtmax" conditions.
dda_rt <- query(dda, "QUERY * WHERE RTMIN = 200 AND RTMAX = 300")
dda_rt## MSn data (Spectra) with 717 spectra in a MsBackendMzR backend:
## msLevel rtime scanIndex
## <integer> <numeric> <integer>
## 1 1 200.053 1554
## 2 1 200.173 1555
## 3 1 200.293 1556
## 4 1 200.413 1557
## 5 1 200.533 1558
## ... ... ... ...
## 713 1 299.176 2266
## 714 1 299.304 2267
## 715 2 299.575 2268
## 716 1 299.713 2269
## 717 2 299.985 2270
## ... 34 more variables/columns.
##
## file(s):
## PestMix1_DDA.mzML
## Processing:
## Filter: select retention time [200..300] on MS level(s) [Fri May 16 19:49:56 2025]Internally, SpectraQL translates the MassQL query into
filter functions that are applied to the Spectra object.
The returned result is now a Spectra object with all
spectra measured between 200 and 300 seconds.
## [1] 200.053 299.985To restrict to MS2 spectra with their precursor m/z matching a
certain value, we can use the "MS2PREC" condition.
Conditions on m/z values support also accuracy specifications. Using
"TOLERANCEMZ" and "TOLERANCEPPM" it is thus
possible to define an absolute and m/z relative acceptable difference.
Below we use such a condition to select MS2 spectra with a precursor m/z
matching 278.093 with a tolerance of 0 and ppm of 20. Since
SpectraQL is case insensitive, we write the full MassQL query
in lower case below.
dda_pmz <- query(
dda,
"query * where ms2prec = 278.093:toleancemz=0:toleranceppm=30")
length(dda_pmz)## [1] 9
dda_pmz## MSn data (Spectra) with 9 spectra in a MsBackendMzR backend:
## msLevel rtime scanIndex
## <integer> <numeric> <integer>
## 1 2 268.587 2041
## 2 2 269.007 2043
## 3 2 269.427 2046
## 4 2 381.709 3058
## 5 2 382.069 3062
## 6 2 382.549 3066
## 7 2 385.879 3105
## 8 2 386.699 3120
## 9 2 387.849 3137
## ... 34 more variables/columns.
##
## file(s):
## PestMix1_DDA.mzML
## Processing:
## Filter: select spectra with precursor m/z matching 278.093 [Fri May 16 19:49:56 2025]We thus selected 9 spectra from the data set with a precursor m/z matching our filter criteria.
precursorMz(dda_pmz)## [1] 278.0936 278.0914 278.0954 278.0921 278.0909 278.0917 278.0913 278.0914
## [9] 278.0918We can obviously also combine the two queries into a single MassQL
query. This time we also omit the "tolerancemz" field as we
set that anyway to a value of 0.
dda_rt <- query(dda,
paste0("query * where rtmin = 200 and rtmax = 300 and ",
"ms2prec = 278.093:toleranceppm = 30"))
dda_rt## MSn data (Spectra) with 3 spectra in a MsBackendMzR backend:
## msLevel rtime scanIndex
## <integer> <numeric> <integer>
## 1 2 268.587 2041
## 2 2 269.007 2043
## 3 2 269.427 2046
## ... 34 more variables/columns.
##
## file(s):
## PestMix1_DDA.mzML
## Processing:
## Filter: select retention time [200..300] on MS level(s) [Fri May 16 19:49:56 2025]
## Filter: select spectra with precursor m/z matching 278.093 [Fri May 16 19:49:56 2025]For conditions "MS2PROD", "MS2PREC" and
"MS1MZ" it is also possible to provide multiple values to
select spectra that match any of the provided m/z values. These values
can be separated by "OR" (or "or"),
"TOLERANCEPPM" and "TOLERANCEMZ" will be
applied to each of these. Below we select all spectra that contain an
(MS1) peak with an m/z matching any of the provided values.
query(dda, "QUERY * WHERE MS1MZ = (123 OR 234.1 OR 300):TOLERANCEMZ=0.05")## MSn data (Spectra) with 2384 spectra in a MsBackendMzR backend:
## msLevel rtime scanIndex
## <integer> <numeric> <integer>
## 1 1 0.471 3
## 2 1 0.591 4
## 3 1 1.191 9
## 4 1 1.311 10
## 5 1 1.551 12
## ... ... ... ...
## 2380 1 899.491 7598
## 2381 1 899.613 7599
## 2382 1 899.747 7600
## 2383 1 899.872 7601
## 2384 1 899.993 7602
## ... 34 more variables/columns.
##
## file(s):
## PestMix1_DDA.mzML
## Processing:
## Filter: select MS level(s) 1 [Fri May 16 19:49:56 2025]Note that using "MS1MZ" will return only MS1 spectra and
"MS2PROD", "MS2MZ" and "MS2PREC"
only MS2 spectra.
Choosing which data to return
Thus far we uses "*" in all queries, which returns the
result as a Spectra object. Alternatively, we can use
"MS1DATA" or "MS2DATA" which will return the
peak data for MS1 or MS2 spectra, respectively. The result is thus not a
Spectra object, but a list of two-column
matrices with the m/z and intensity values of all selected spectra.
Below we use this to extract the peaks data for all MS2 spectra measured
between 200 and 300 seconds.
## [1] 216
head(pks[[1L]])## mz intensity
## [1,] 54.93746 0.021841669
## [2,] 55.93555 0.173691928
## [3,] 70.94262 0.011819169
## [4,] 71.71149 0.017824525
## [5,] 71.72899 0.005942247
## [6,] 71.93047 0.834873378We can in addition also use functions to extract only specific data
from the result. "scansum" would for example return the sum
of intensities per spectra and would thus allow to extract a total ion
chromatogram:
tic <- query(dda, "query scansum(ms1data)")
plot(tic, type = "l", xlab = "scan index")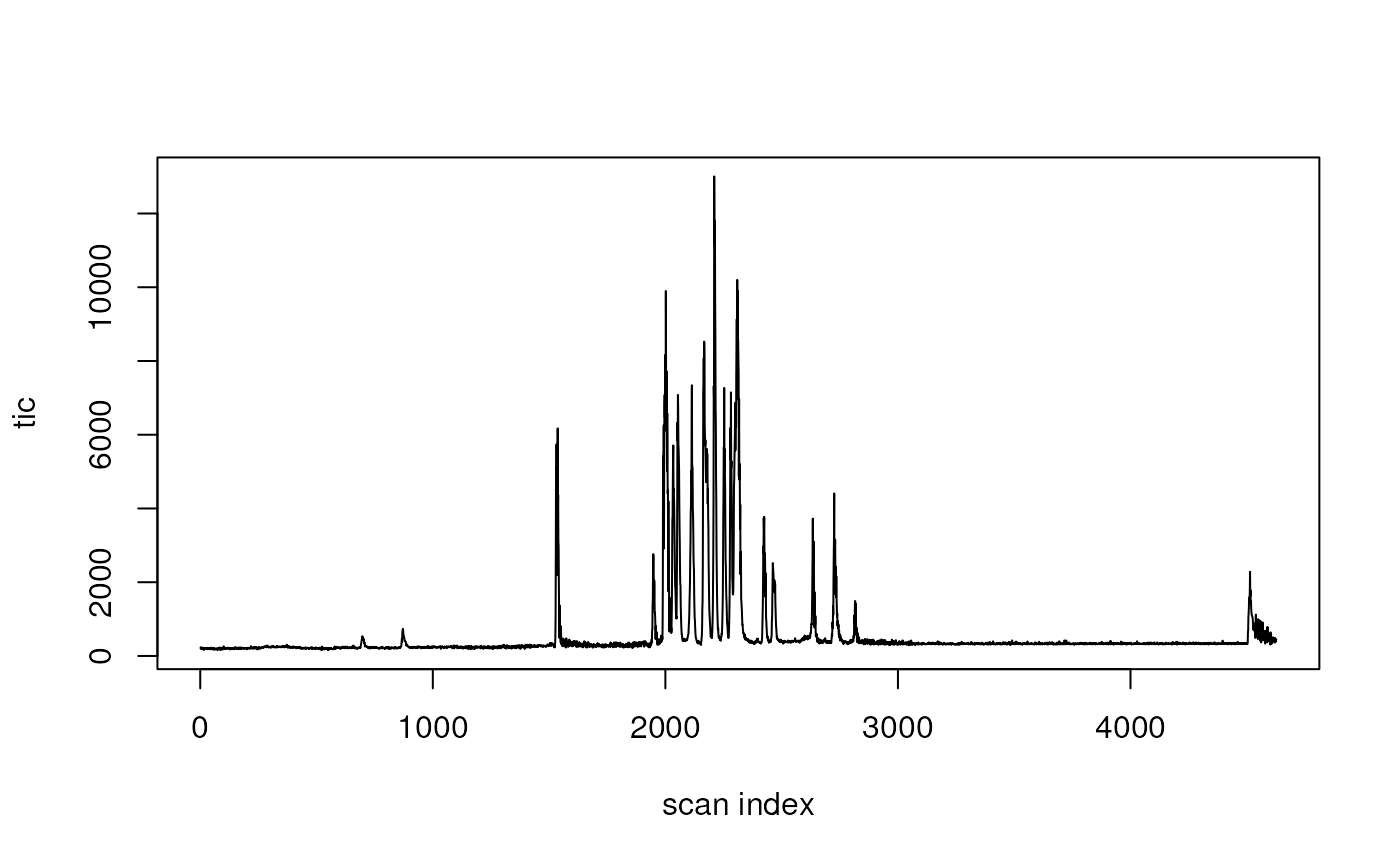
Please note that the x-axis does not represent the retention times but the scan indices.
The function "scaninfo" extracts all information from
the selected spectra. For SpectraQL this means that the result
from spectraData() are returned:
si <- query(dda, "query scaninfo(*) where rtmin = 300 and rtmax = 500")
si## DataFrame with 2236 rows and 35 columns
## msLevel rtime acquisitionNum scanIndex dataStorage
## <integer> <numeric> <integer> <integer> <character>
## 1 1 300.123 2271 2271 /__w/_temp/Library/m..
## 2 2 300.395 2272 2272 /__w/_temp/Library/m..
## 3 1 300.533 2273 2273 /__w/_temp/Library/m..
## 4 1 300.654 2274 2274 /__w/_temp/Library/m..
## 5 1 300.775 2275 2275 /__w/_temp/Library/m..
## ... ... ... ... ... ...
## 2232 2 499.391 4502 4502 /__w/_temp/Library/m..
## 2233 1 499.529 4503 4503 /__w/_temp/Library/m..
## 2234 2 499.691 4504 4504 /__w/_temp/Library/m..
## 2235 2 499.811 4505 4505 /__w/_temp/Library/m..
## 2236 1 499.948 4506 4506 /__w/_temp/Library/m..
## dataOrigin centroided smoothed polarity precScanNum
## <character> <logical> <logical> <integer> <integer>
## 1 /__w/_temp/Library/m.. TRUE NA 1 NA
## 2 /__w/_temp/Library/m.. TRUE NA 1 NA
## 3 /__w/_temp/Library/m.. TRUE NA 1 NA
## 4 /__w/_temp/Library/m.. TRUE NA 1 NA
## 5 /__w/_temp/Library/m.. TRUE NA 1 NA
## ... ... ... ... ... ...
## 2232 /__w/_temp/Library/m.. TRUE NA 1 NA
## 2233 /__w/_temp/Library/m.. TRUE NA 1 NA
## 2234 /__w/_temp/Library/m.. TRUE NA 1 NA
## 2235 /__w/_temp/Library/m.. TRUE NA 1 NA
## 2236 /__w/_temp/Library/m.. TRUE NA 1 NA
## precursorMz precursorIntensity precursorCharge collisionEnergy
## <numeric> <numeric> <integer> <numeric>
## 1 NA NA NA NA
## 2 320.106 0 0 0
## 3 NA NA NA NA
## 4 NA NA NA NA
## 5 NA NA NA NA
## ... ... ... ... ...
## 2232 252.995 0 0 0
## 2233 NA NA NA NA
## 2234 173.079 0 0 0
## 2235 368.182 0 0 0
## 2236 NA NA NA NA
## isolationWindowLowerMz isolationWindowTargetMz isolationWindowUpperMz
## <numeric> <numeric> <numeric>
## 1 NA NA NA
## 2 319.606 320.106 320.606
## 3 NA NA NA
## 4 NA NA NA
## 5 NA NA NA
## ... ... ... ...
## 2232 252.495 252.995 253.495
## 2233 NA NA NA
## 2234 172.579 173.079 173.579
## 2235 367.682 368.182 368.682
## 2236 NA NA NA
## peaksCount totIonCurrent basePeakMZ basePeakIntensity ionisationEnergy
## <integer> <numeric> <numeric> <numeric> <numeric>
## 1 327 236402 54.0089 44069 0
## 2 52 5440 233.0023 288 0
## 3 348 221394 54.0089 39904 0
## 4 198 211449 54.0089 43642 0
## 5 377 248240 54.0099 32575 0
## ... ... ... ... ... ...
## 2232 2 1025 98.9837 32 0
## 2233 370 307324 54.0088 24136 0
## 2234 10 1360 55.0552 42 0
## 2235 4 1103 368.1747 42 0
## 2236 426 318309 54.0088 25913 0
## lowMZ highMZ mergedScan mergedResultScanNum
## <numeric> <numeric> <integer> <integer>
## 1 0 0 NA NA
## 2 0 0 NA NA
## 3 0 0 NA NA
## 4 0 0 NA NA
## 5 0 0 NA NA
## ... ... ... ... ...
## 2232 0 0 NA NA
## 2233 0 0 NA NA
## 2234 0 0 NA NA
## 2235 0 0 NA NA
## 2236 0 0 NA NA
## mergedResultStartScanNum mergedResultEndScanNum injectionTime filterString
## <integer> <integer> <numeric> <character>
## 1 NA NA 0 NA
## 2 NA NA 0 NA
## 3 NA NA 0 NA
## 4 NA NA 0 NA
## 5 NA NA 0 NA
## ... ... ... ... ...
## 2232 NA NA 0 NA
## 2233 NA NA 0 NA
## 2234 NA NA 0 NA
## 2235 NA NA 0 NA
## 2236 NA NA 0 NA
## spectrumId ionMobilityDriftTime scanWindowLowerLimit
## <character> <numeric> <numeric>
## 1 sample=1 period=1 cy.. NA 50
## 2 sample=1 period=1 cy.. NA 50
## 3 sample=1 period=1 cy.. NA 50
## 4 sample=1 period=1 cy.. NA 50
## 5 sample=1 period=1 cy.. NA 50
## ... ... ... ...
## 2232 sample=1 period=1 cy.. NA 50
## 2233 sample=1 period=1 cy.. NA 50
## 2234 sample=1 period=1 cy.. NA 50
## 2235 sample=1 period=1 cy.. NA 50
## 2236 sample=1 period=1 cy.. NA 50
## scanWindowUpperLimit electronBeamEnergy
## <numeric> <numeric>
## 1 2000 NA
## 2 2000 NA
## 3 2000 NA
## 4 2000 NA
## 5 2000 NA
## ... ... ...
## 2232 2000 NA
## 2233 2000 NA
## 2234 2000 NA
## 2235 2000 NA
## 2236 2000 NAFiltering peaks within spectra
In contrast to the condition statements
("WHERE"), "FILTER" allows to filter the peak
data within spectra. We can for example filter all spectra keeping only
peaks with a certain m/z. Below we filter the MS1 data keeping only
peaks with an m/z value matching 219.095 with a tolerance of 0.01 and
extract the ion count for that.
ic <- query(
dda, "query scansum(ms1data) filter ms1mz = 219.095:tolerancemz=0.1")
plot(ic, type = "l", xlab = "scan index")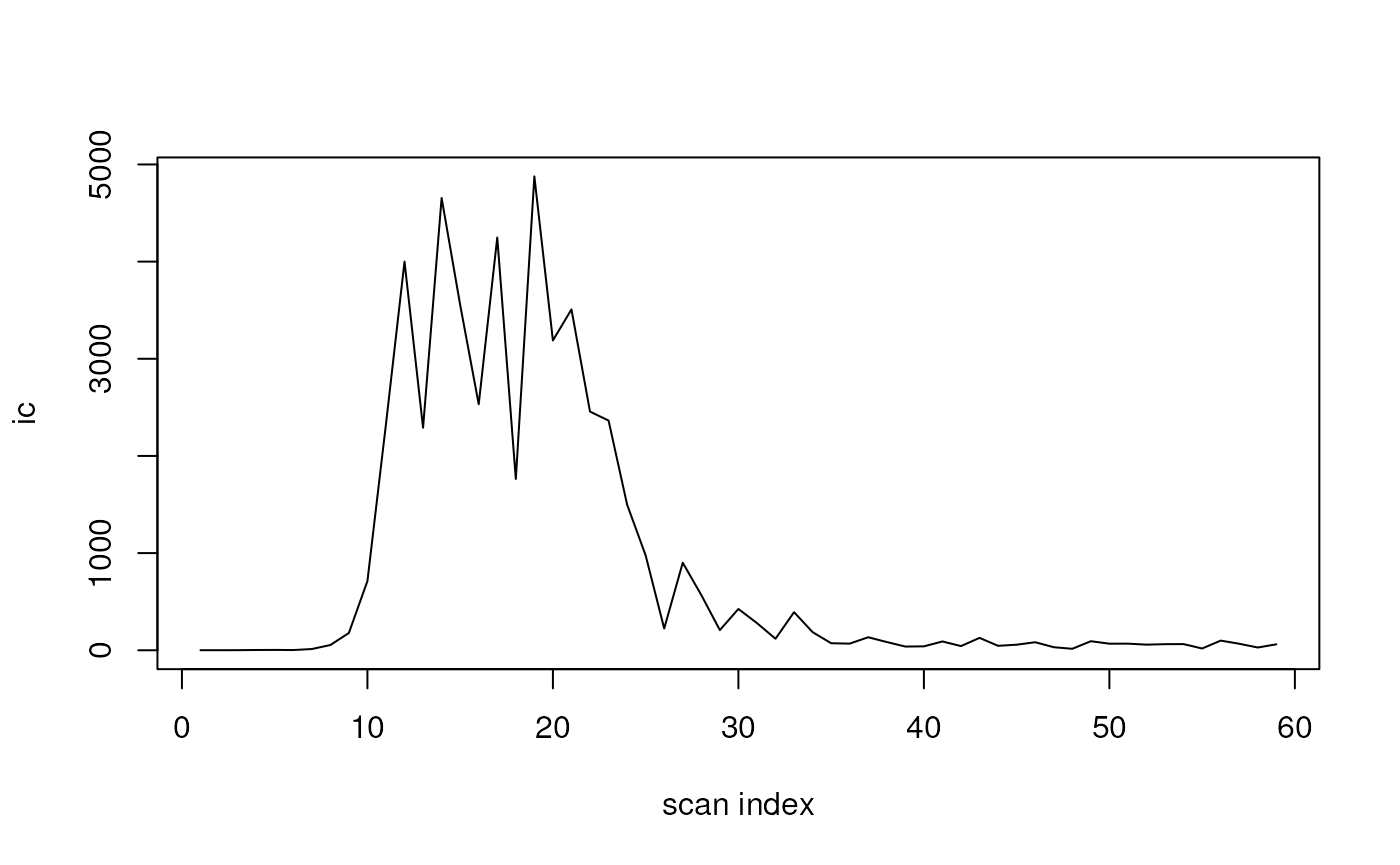
We can also combine that with "QUERY" to restrict to a
certain retention time range to generate an extracted ion chromatogram
(XIC).
ic <- query(
dda,
paste0("query scansum(ms1data) where rtmin = 235 and rtmax = 250",
" filter ms1mz = 219.095:tolerancemz=0.1"))
plot(ic, type = "l", xlab = "scan index")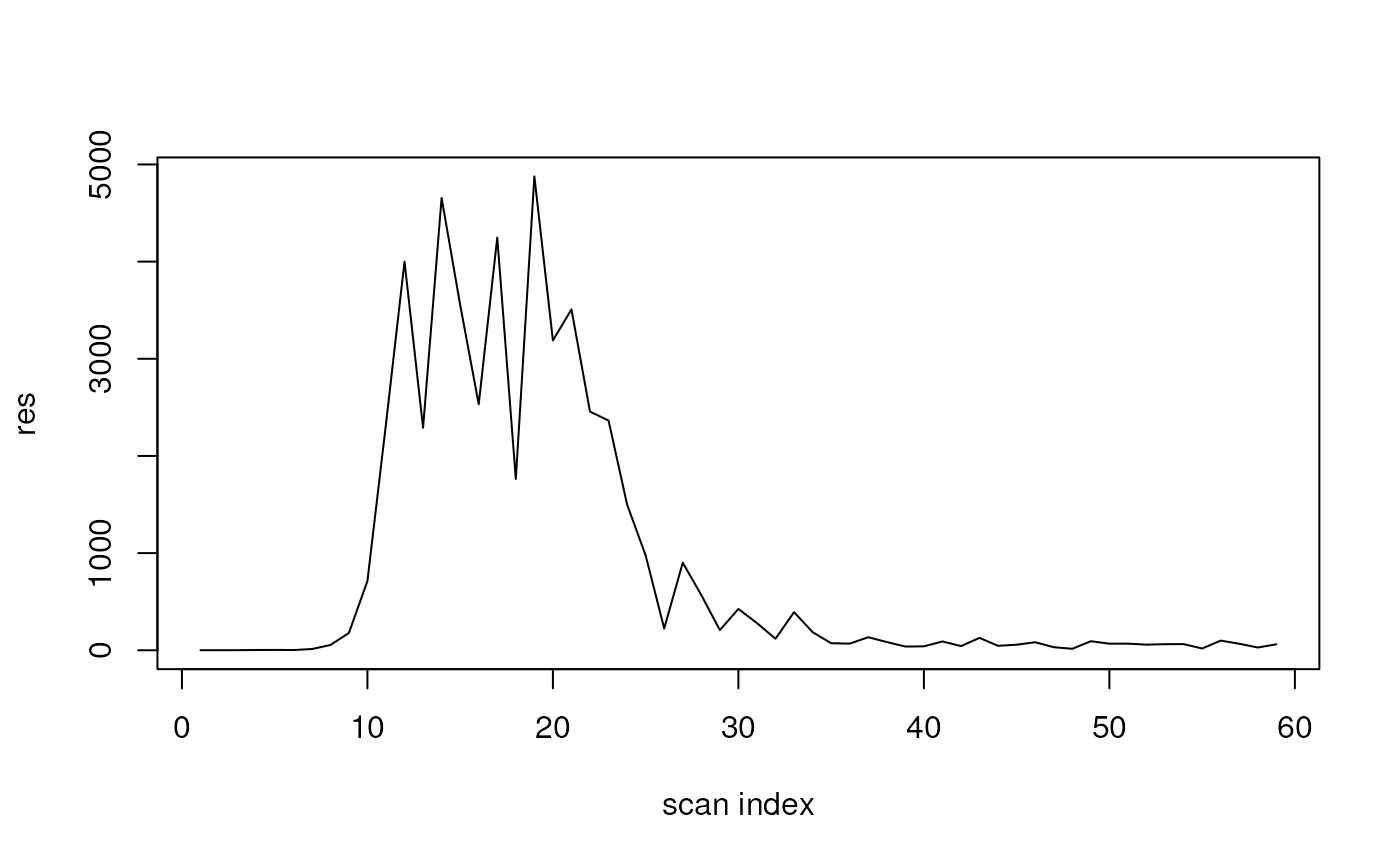
Internally, SpectraQL’s function translates the MassQL query into the following combination of function calls:
res <- dda |>
filterMsLevel(1L) |>
filterRt(c(235, 250)) |>
filterMzValues(219.095, tolerance = 0.1) |>
ionCount()
plot(res, type = "l", xlab = "scan index")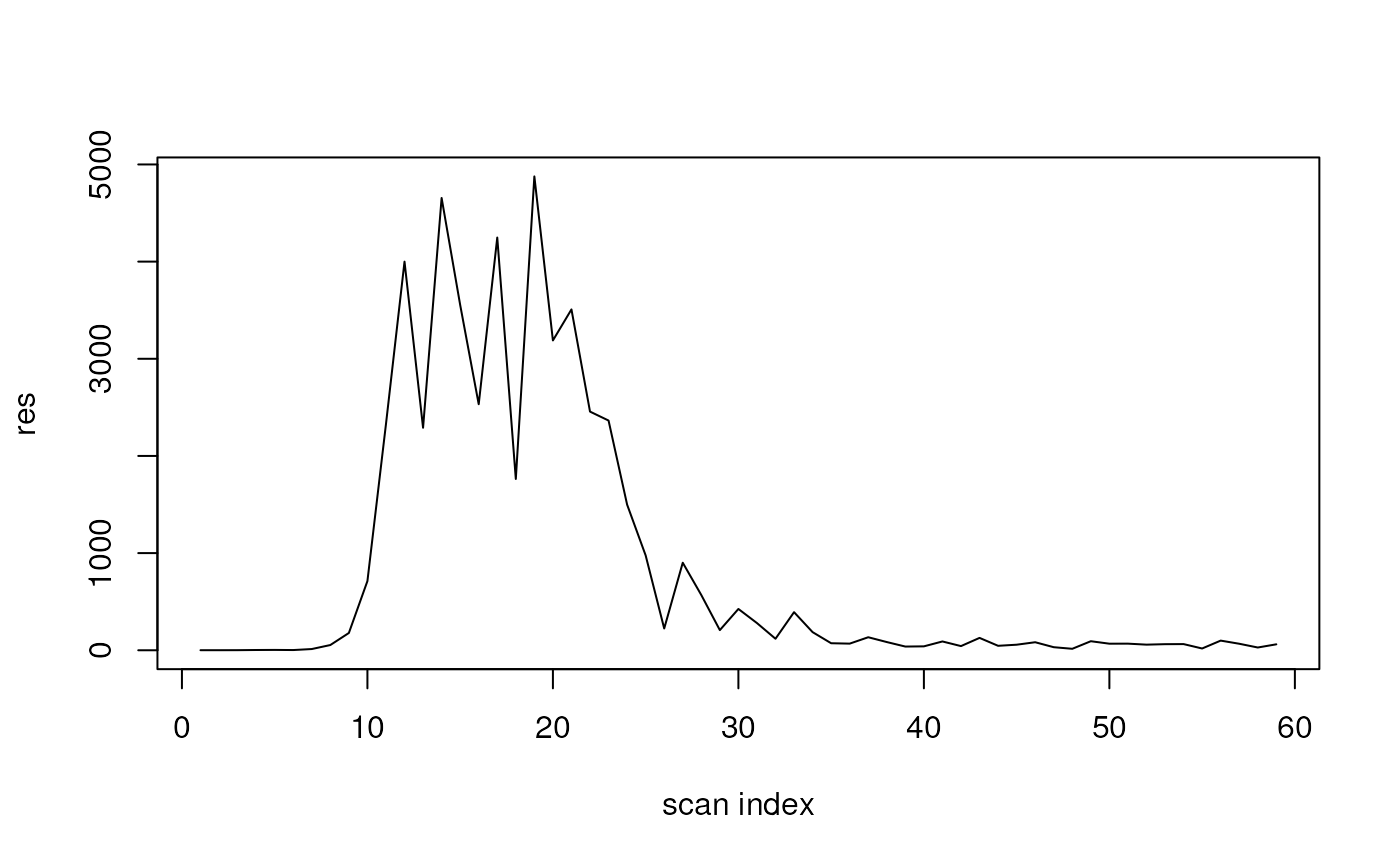
Session information
## R version 4.5.0 (2025-04-11)
## Platform: x86_64-pc-linux-gnu
## Running under: Ubuntu 24.04.2 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
## LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## time zone: UTC
## tzcode source: system (glibc)
##
## attached base packages:
## [1] stats4 stats graphics grDevices utils datasets methods
## [8] base
##
## other attached packages:
## [1] msdata_0.49.0 SpectraQL_1.3.1 ProtGenerics_1.39.2
## [4] Spectra_1.19.0 BiocParallel_1.43.0 S4Vectors_0.47.0
## [7] BiocGenerics_0.55.0 generics_0.1.4 BiocStyle_2.37.0
##
## loaded via a namespace (and not attached):
## [1] jsonlite_2.0.0 compiler_4.5.0 BiocManager_1.30.25
## [4] Rcpp_1.0.14 Biobase_2.69.0 parallel_4.5.0
## [7] cluster_2.1.8.1 jquerylib_0.1.4 systemfonts_1.2.3
## [10] IRanges_2.43.0 textshaping_1.0.1 yaml_2.3.10
## [13] fastmap_1.2.0 R6_2.6.1 knitr_1.50
## [16] htmlwidgets_1.6.4 MASS_7.3-65 bookdown_0.43
## [19] desc_1.4.3 bslib_0.9.0 rlang_1.1.6
## [22] cachem_1.1.0 xfun_0.52 fs_1.6.6
## [25] MsCoreUtils_1.21.0 sass_0.4.10 cli_3.6.5
## [28] pkgdown_2.1.2.9000 ncdf4_1.24 digest_0.6.37
## [31] mzR_2.43.0 MetaboCoreUtils_1.17.1 lifecycle_1.0.4
## [34] clue_0.3-66 evaluate_1.0.3 codetools_0.2-20
## [37] ragg_1.4.0 rmarkdown_2.29 tools_4.5.0
## [40] htmltools_0.5.8.1