Creates a list of annotations based on calculateFragments results.
labelFragments(x, tolerance = 0, ppm = 20, what = c("ion", "mz"), ...)
addFragments(x, tolerance = 0, ppm = 20, ...)Arguments
- x
An instance of class
Spectraof length 1, containing a spectra variable"sequence"with acharacter(1)representing a valid peptide sequence.- tolerance
absolute acceptable difference of m/z values for peaks to be considered matching (see
MsCoreUtils::common()for more details).- ppm
m/z relative acceptable difference (in ppm) for peaks to be considered matching (see
MsCoreUtils::common()for more details).- what
character(1), one of"ion"(default) or"mz", defining whether labels should be fragment ions, , or their m/z values. If the latter, then the m/z values are named with the ion labels.- ...
additional parameters (except
verbose) passed tocalculateFragments()to calculate fragment m/z values to be added to the spectra inx.
Value
Return a list() of character() with fragment ion labels. The
elements are named after the peptide they belong to (variable
modifications included).
Details
addFragments is deprecated and will be made defunct; use labelFragments
instead.
Examples
library("Spectra")
#> Loading required package: BiocParallel
sp <- DataFrame(msLevel = 2L, rtime = 2345, sequence = "SIGFEGDSIGR")
sp$mz <- list(c(100.048614501953, 110.069030761719, 112.085876464844,
117.112571716309, 158.089569091797, 163.114898681641,
175.117172241211, 177.098587036133, 214.127075195312,
232.137542724609, 233.140335083008, 259.938415527344,
260.084167480469, 277.111572265625, 282.680786132812,
284.079437255859, 291.208282470703, 315.422576904297,
317.22509765625, 327.2060546875, 362.211944580078,
402.235290527344, 433.255004882812, 529.265991210938,
549.305236816406, 593.217041015625, 594.595092773438,
609.848327636719, 631.819702148438, 632.324035644531,
632.804931640625, 640.8193359375, 641.309936523438,
641.82568359375, 678.357238769531, 679.346252441406,
688.291259765625, 735.358947753906, 851.384033203125,
880.414001464844, 881.40185546875, 919.406433105469,
938.445861816406, 1022.56658935547, 1050.50415039062,
1059.82800292969, 1107.52734375, 1138.521484375,
1147.51538085938, 1226.056640625))
sp$intensity <- list(c(83143.03, 65473.8, 192735.53, 3649178.5,
379537.81, 89117.58, 922802.69, 61190.44,
281353.22, 2984798.75, 111935.03, 42512.57,
117443.59, 60773.67, 39108.15, 55350.43,
209952.97, 37001.18, 439515.53, 139584.47,
46842.71, 1015457.44, 419382.31, 63378.77,
444406.66, 58426.91, 46007.71, 58711.72,
80675.59, 312799.97, 134451.72, 151969.72,
3215457.75, 1961975, 395735.62, 71002.98,
69405.73, 136619.47, 166158.69, 682329.75,
239964.69, 242025.44, 1338597.62, 50118.02,
1708093.12, 43119.03, 97048.02, 2668231.75,
83310.2, 40705.72))
sp <- Spectra(sp)
## The fragment ion labels
labelFragments(sp)
#> $SIGFEGDSIGR
#> [1] NA NA NA NA NA NA "y1" NA "y2_" "y2" NA NA
#> [13] NA NA NA NA NA NA NA NA NA NA NA "y5_"
#> [25] NA NA NA NA NA NA NA NA NA NA NA NA
#> [37] NA NA NA "y8" NA NA NA NA "y10" NA NA NA
#> [49] NA NA
#>
#> attr(,"group")
#> [1] 1
## The fragment mz labels
labelFragments(sp, what = "mz")
#> $SIGFEGDSIGR
#> [1] NA NA NA NA NA NA 175.1190
#> [8] NA 214.1298 232.1404 NA NA NA NA
#> [15] NA NA NA NA NA NA NA
#> [22] NA NA 529.2729 NA NA NA NA
#> [29] NA NA NA NA NA NA NA
#> [36] NA NA NA NA 880.4159 NA NA
#> [43] NA NA 1050.5214 NA NA NA NA
#> [50] NA
#>
#> attr(,"group")
#> [1] 1
## Call additional parameters sur as variable modifications to calculateFragments
labelFragments(sp, type = c("a", "b", "x", "y"), variable_modifications = c(R = 5))
#> $SIGFEGDSIGR
#> [1] NA NA NA NA NA NA "y1" NA "y2_" "y2" NA NA
#> [13] NA NA NA NA NA NA NA NA NA NA NA "y5_"
#> [25] NA NA NA NA NA NA NA NA NA NA NA NA
#> [37] NA NA NA "y8" NA NA NA NA "y10" NA NA NA
#> [49] NA NA
#>
#> $`SIGFEGDSIGR[5]`
#> [1] NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA
#> [26] NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA
#>
#> attr(,"group")
#> [1] 1 1
## Annotate the spectum with the fragment labels
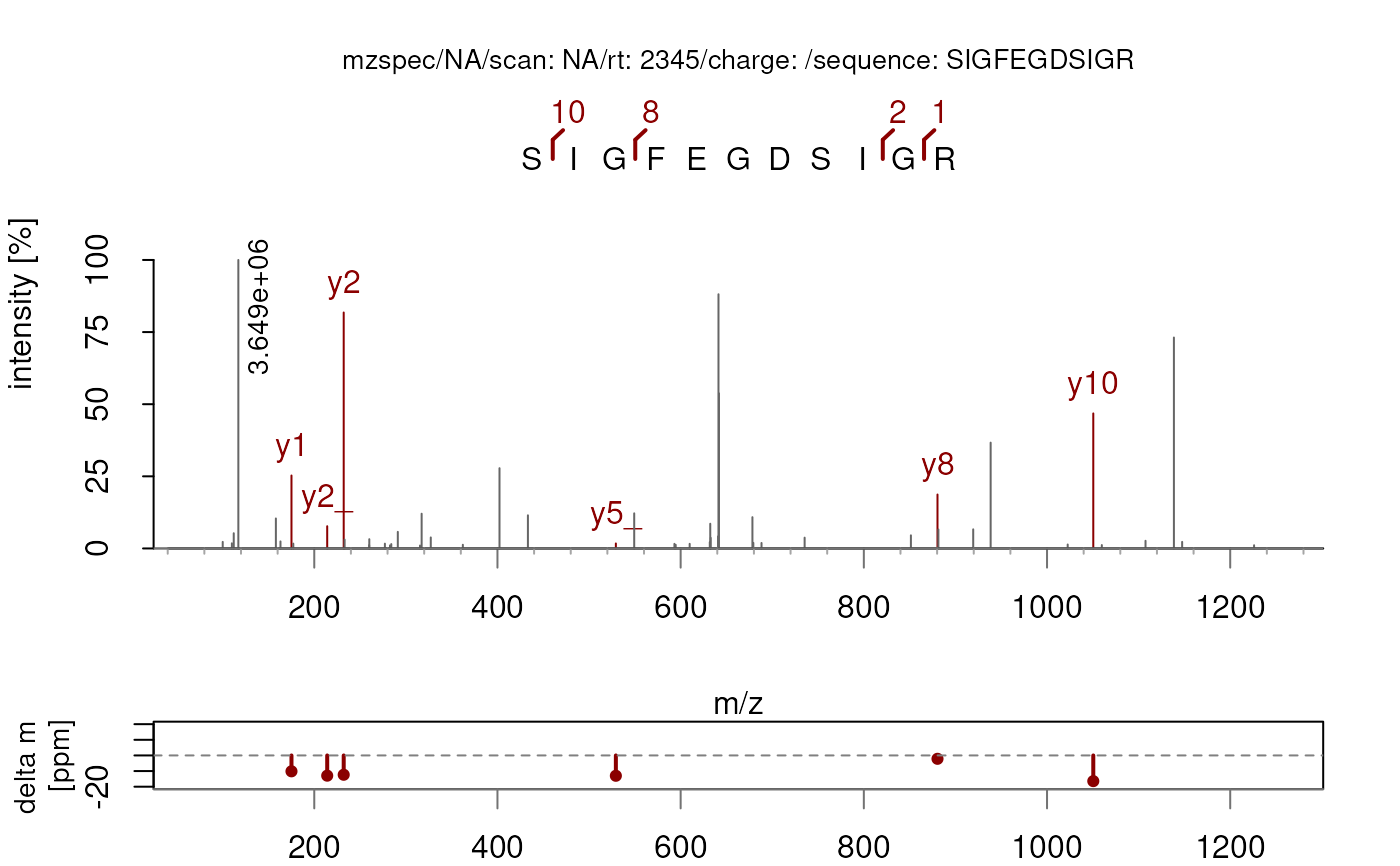
plotSpectra(sp, labels = labelFragments, labelPos = 3)
 ## By default used in `plotSpectraPTM()`.
plotSpectraPTM(sp)
## By default used in `plotSpectraPTM()`.
plotSpectraPTM(sp)