1.3 Dependences and connectivity of metabolomics packages
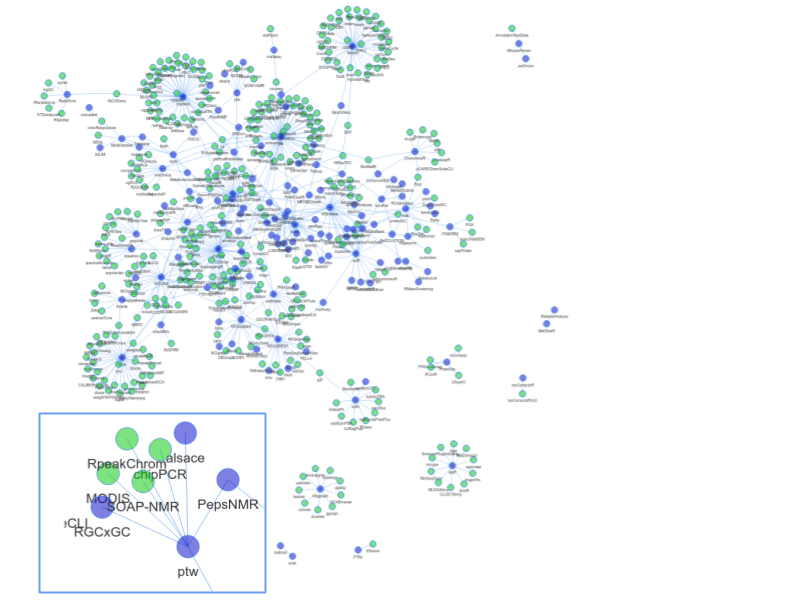
Figure 2: Dependency network of R packages. Shown in blue are packages mentioned in the review. Edges connect to packages that depend on another package, as long as they are in CRAN or BioC. Green nodes correspond to packages in CRAN or BioC not covered in the review. The inset shows the neighbourhood of the ptw package. Not shown are 1) infrastructure packages, e.g. rJava, Rcpp 2) packages from the review without reverse dependencies and 3) data packages. Some packages from the review are not in current versions of CRAN or BioC. An interactive version of this figure is also available online (rformassspectrometry.github.io/metaRbolomics-book, Appendix 2) and as supplemental file 2.
Code reuse and object inheritance can be a sign for a well-connected and interacting community. At the useR!2015 and JSM2015 conferences, A. de Vries and J. Rickert (both Microsoft, London, UK) showed the analysis of the CRAN and BioC dependency network structure [23–25]. Compared to CRAN, BioC packages had a higher connectivity: “It seems that the Bioconductor policy encourages package authors to reuse existing material and write packages that work better together”. We repeated such an analysis [26] with the packages mentioned in this review and created a network of reverse dependencies (i.e., the set of packages that depend on these metabolomics related packages in BioC or CRAN). The resulting network is shown in Figure 2.