Connected components
Source:R/ConnectedComponent-class.R, R/adjacencyMatrix-accessors.R
ConnectedComponents.RdConnected components are a useful representation when exploring identification data. They represent the relation between proteins (the connected components) and how they form groups of proteins as defined by shared peptides.
Connected components are stored as ConnectedComponents objects
that can be generated using the ConnectedComponents()
function.
ConnectedComponents(object, ...)
ccMatrix(x)
connectedComponents(x, i, simplify = TRUE)
# S4 method for class 'ConnectedComponents'
length(x)
# S4 method for class 'ConnectedComponents'
dims(x)
# S4 method for class 'ConnectedComponents'
ncols(x)
# S4 method for class 'ConnectedComponents'
nrows(x)
# S4 method for class 'ConnectedComponents,integer,ANY,ANY'
x[i, j, ..., drop = FALSE]
# S4 method for class 'ConnectedComponents,logical,ANY,ANY'
x[i, j, ..., drop = FALSE]
# S4 method for class 'ConnectedComponents,numeric,ANY,ANY'
x[i, j, ..., drop = FALSE]
prioritiseConnectedComponents(x)
prioritizeConnectedComponents(x)
# S4 method for class 'ConnectedComponents'
adjacencyMatrix(object)Arguments
- object
For the
ConnectedComponentsclass constructor, either a sparse adjacency matrix of classMatrixor an instance of classPSM.- ...
Additional arguments passed to
makeAdjacencyMatrix()whenobjectis of classPSM().- x
An object of class
ConnectedComponents.- i
numeric(),integer()orlogical()to subset theConnectedComponentsinstance. If alogical(), it must be of same length as the object is subsets.- simplify
logical(1)ifTRUE(default), the output is simplified to sparse matrix ifiwas of length 1, otherwise aListis returned. Always aListifFALSE.- j
ignored
- drop
ignore
Value
The ConnectedComponents() constructor returns an
instance of class ConnectedComponents. The Creating and
manipulating objects section describes the return values of
the functions that manipulate ConnectedComponents objects.
Slots
adjMatrixThe sparse adjacency matrix (class
Matrix) of dimension p peptides by m proteins that was used to generate the object.ccMatrixThe sparse connected components matrix (class
Matrix) of dimension m by m proteins.adjMatricesA
Listcontaining adjacency matrices of each connected components.
Creating and manipulating objects
Instances of the class are created with the
ConnectedComponent()constructor from aPSM()object or directly from a sparse adjacency matrix of classMatrix. Note that if using the latter, the rows and columns must be named.The sparse peptide-by-protein adjacency matrix is stored in the
ConnectedComponentinstance and can be accessed with theadjacencyMatrix()function.The protein-by-protein connected components sparse matrix of object
xcan be accessed with theccMatrix(x)function.The number of connected components of object
xcan be retrieved withlength(x).The size of the connected components of object
x, i.e the number of proteins in each component, can be retrieved withncols(x). The number of peptides defining the connected components can be retrieved withnrows(x). Both can be accessed withdims(x).The
connectedComponents(x, i, simplify = TRUE)function returns the peptide-by-protein sparse adjacency matrix (orListof matrices, iflength(i) > 1), i.e. the subset of the adjacency matrix defined by the proteins in connected component(s)i.iis the numeric index (between 1 andlength(x)) of the connected connected. If simplify isTRUE(default), then a matrix is returned instead of aListof matrices of length 1. If set toFALSE, aListis always returned, irrespective of its length.To help with the exploration of individual connected Components, the
prioritiseConnectedComponents()function will take an instance ofConnectedComponentsand return adata.framewhere the component indices are ordered based on their potential to clean up/flag some peptides and split protein groups in small groups or individual proteins, or simply explore them. The prioritisation is based on a set of metrics computed from the component's adjacency matrix, including its dimensions, row and col sums maxima and minima, its sparsity and the number of communities and their modularity that quantifies how well the communities separate (seeigraph::modularity(). Note that trivial components, i.e. those composed of a single peptide and protein are excluded from the prioritised results. Thisdata.frameis ideally suited for a principal component analysis (using for instanceprcomp()) for further inspection for component visualisation withplotAdjacencyMatrix().
Examples
## --------------------------------
## From an adjacency matrix
## --------------------------------
library(Matrix)
#>
#> Attaching package: ‘Matrix’
#> The following object is masked from ‘package:S4Vectors’:
#>
#> expand
adj <- sparseMatrix(i = c(1, 2, 3, 3, 4, 4, 5),
j = c(1, 2, 3, 4, 3, 4, 5),
x = 1,
dimnames = list(paste0("Pep", 1:5),
paste0("Prot", 1:5)))
adj
#> 5 x 5 sparse Matrix of class "dgCMatrix"
#> Prot1 Prot2 Prot3 Prot4 Prot5
#> Pep1 1 . . . .
#> Pep2 . 1 . . .
#> Pep3 . . 1 1 .
#> Pep4 . . 1 1 .
#> Pep5 . . . . 1
cc <- ConnectedComponents(adj)
cc
#> An instance of class ConnectedComponents
#> Number of proteins: 5
#> Number of components: 4
#> Number of components [peptide x proteins]:
#> 3[1 x 1] 0[1 x n] 0[n x 1] 1[n x n]
length(cc)
#> [1] 4
ncols(cc)
#> [1] 1 1 2 1
adjacencyMatrix(cc) ## same as adj above
#> 5 x 5 sparse Matrix of class "dgCMatrix"
#> Prot1 Prot2 Prot3 Prot4 Prot5
#> Pep1 1 . . . .
#> Pep2 . 1 . . .
#> Pep3 . . 1 1 .
#> Pep4 . . 1 1 .
#> Pep5 . . . . 1
ccMatrix(cc)
#> 5 x 5 sparse Matrix of class "dsCMatrix"
#> Prot1 Prot2 Prot3 Prot4 Prot5
#> Prot1 1 . . . .
#> Prot2 . 1 . . .
#> Prot3 . . 2 2 .
#> Prot4 . . 2 2 .
#> Prot5 . . . . 1
connectedComponents(cc)
#> List of length 4
connectedComponents(cc, 3) ## a singel matrix
#> 2 x 2 sparse Matrix of class "dgCMatrix"
#> Prot3 Prot4
#> Pep3 1 1
#> Pep4 1 1
connectedComponents(cc, 1:2) ## a List
#> List of length 2
## --------------------------------
## From an PSM object
## --------------------------------
f <- msdata::ident(full.names = TRUE, pattern = "TMT")
f
#> [1] "/__w/_temp/Library/msdata/ident/TMT_Erwinia_1uLSike_Top10HCD_isol2_45stepped_60min_01-20141210.mzid"
psm <- PSM(f) |>
filterPsmDecoy() |>
filterPsmRank()
#> Loading required namespace: mzR
#> Removed 2896 decoy hits.
#> Removed 155 PSMs with rank > 1.
cc <- ConnectedComponents(psm)
cc
#> An instance of class ConnectedComponents
#> Number of proteins: 1504
#> Number of components: 1476
#> Number of components [peptide x proteins]:
#> 954[1 x 1] 7[1 x n] 501[n x 1] 14[n x n]
length(cc)
#> [1] 1476
table(ncols(cc))
#>
#> 1 2 3 4
#> 1455 17 1 3
(i <- which(ncols(cc) == 4))
#> [1] 689 1079 1082
ccomp <- connectedComponents(cc, i)
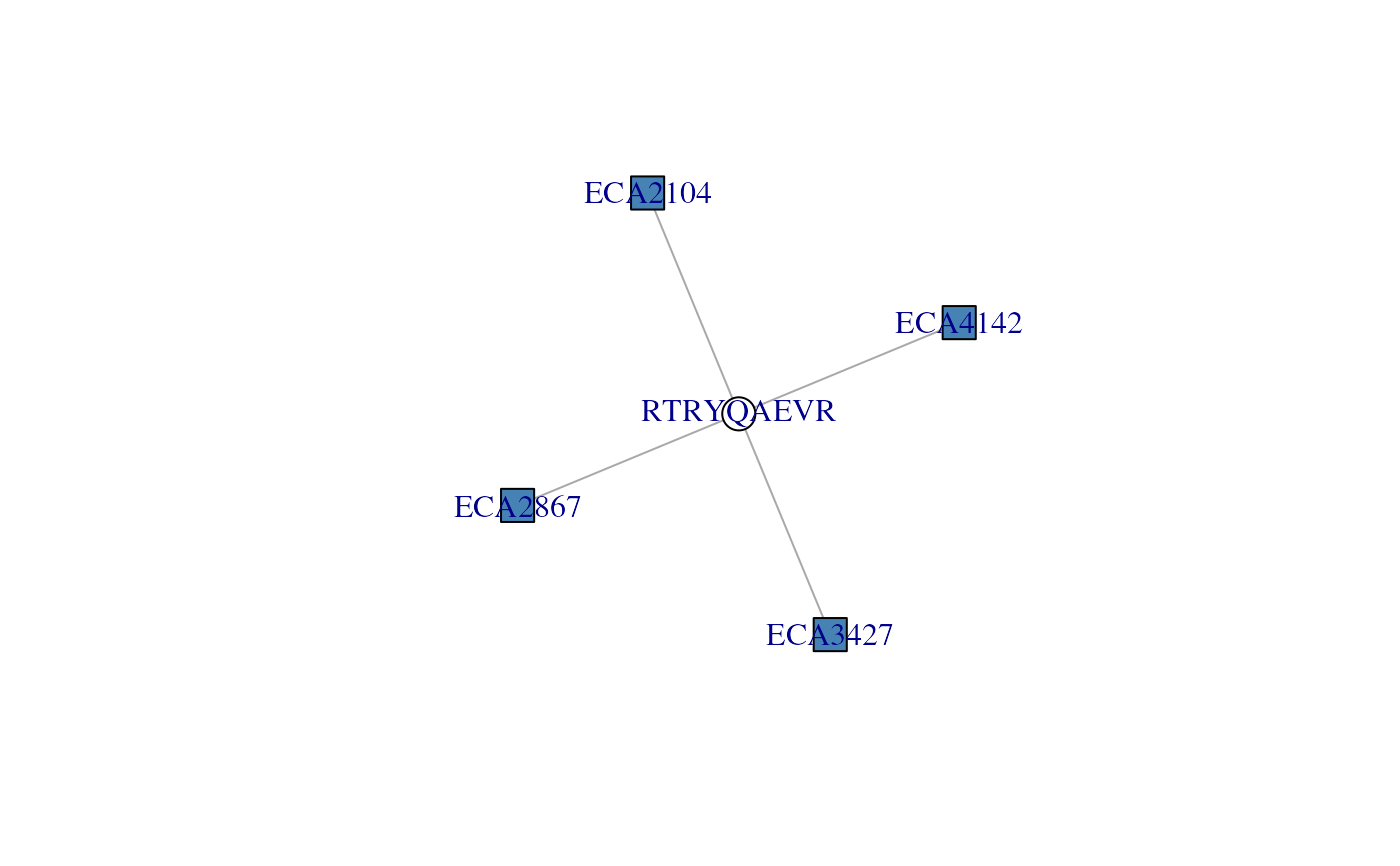
## A group of 4 proteins that all share peptide RTRYQAEVR
ccomp[[1]]
#> 1 x 4 sparse Matrix of class "dgCMatrix"
#> ECA2104 ECA2867 ECA3427 ECA4142
#> RTRYQAEVR 1 1 1 1
## Visualise the adjacency matrix - here, we see how the single
## peptides (white node) 'unites' the four proteins (blue nodes)
plotAdjacencyMatrix(ccomp[[1]])
#> Warning: vertex attribute color contains NAs. Replacing with default value 1
#> Warning: vertex attribute color contains NAs. Replacing with default value 1
#> Warning: vertex attribute color contains NAs. Replacing with default value 1
#> Warning: vertex attribute color contains NAs. Replacing with default value 1
#> Warning: vertex attribute color contains NAs. Replacing with default value 1
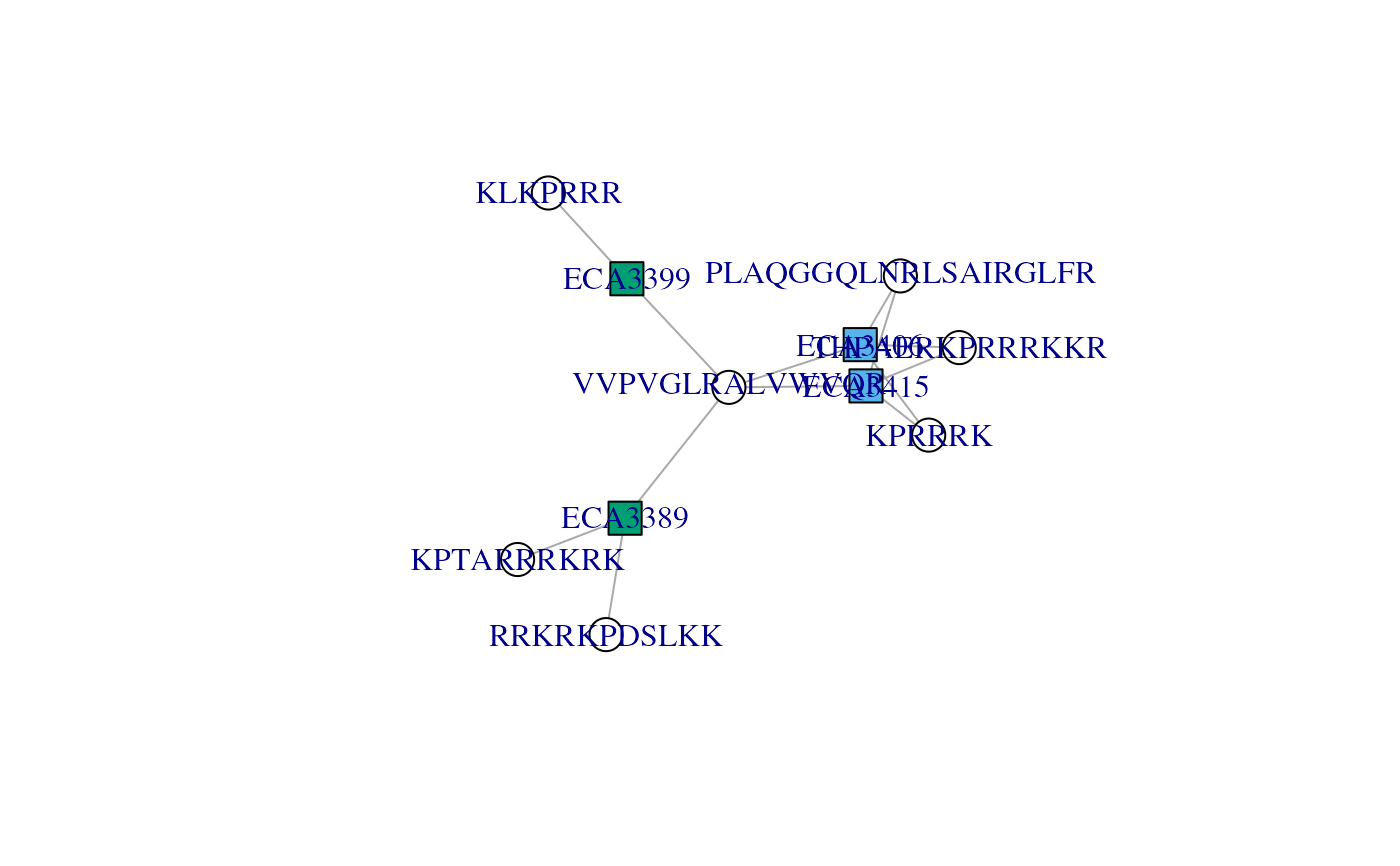
 ## A group of 4 proteins formed by 7 peptides: THPAERKPRRRKKR is
## found in the two first proteins, KPTARRRKRK was found twice in
## ECA3389, VVPVGLRALVWVQR was found in all 4 proteins, KLKPRRR
## is specific to ECA3399, ...
ccomp[[3]]
#> 7 x 4 sparse Matrix of class "dgCMatrix"
#> ECA3406 ECA3415 ECA3389 ECA3399
#> THPAERKPRRRKKR 1 1 . .
#> KPTARRRKRK . . 2 .
#> PLAQGGQLNRLSAIRGLFR 1 1 . .
#> RRKRKPDSLKK . . 1 .
#> KPRRRK 1 1 . .
#> VVPVGLRALVWVQR 1 1 1 1
#> KLKPRRR . . . 1
## See how VVPVGLRALVWVQR is shared by ECA3406 ECA3415 ECA3389 and
## links the three other componennts, namely ECA3399, ECA3389 and
## (ECA3415, ECA3406). Filtering that peptide out would split that
## protein group in three.
plotAdjacencyMatrix(ccomp[[3]])
#> Warning: vertex attribute color contains NAs. Replacing with default value 1
#> Warning: vertex attribute color contains NAs. Replacing with default value 1
#> Warning: vertex attribute color contains NAs. Replacing with default value 1
#> Warning: vertex attribute color contains NAs. Replacing with default value 1
#> Warning: vertex attribute color contains NAs. Replacing with default value 1
#> Warning: vertex attribute color contains NAs. Replacing with default value 1
#> Warning: vertex attribute color contains NAs. Replacing with default value 1
#> Warning: vertex attribute color contains NAs. Replacing with default value 1
#> Warning: vertex attribute color contains NAs. Replacing with default value 1
#> Warning: vertex attribute color contains NAs. Replacing with default value 1
#> Warning: vertex attribute color contains NAs. Replacing with default value 1
## A group of 4 proteins formed by 7 peptides: THPAERKPRRRKKR is
## found in the two first proteins, KPTARRRKRK was found twice in
## ECA3389, VVPVGLRALVWVQR was found in all 4 proteins, KLKPRRR
## is specific to ECA3399, ...
ccomp[[3]]
#> 7 x 4 sparse Matrix of class "dgCMatrix"
#> ECA3406 ECA3415 ECA3389 ECA3399
#> THPAERKPRRRKKR 1 1 . .
#> KPTARRRKRK . . 2 .
#> PLAQGGQLNRLSAIRGLFR 1 1 . .
#> RRKRKPDSLKK . . 1 .
#> KPRRRK 1 1 . .
#> VVPVGLRALVWVQR 1 1 1 1
#> KLKPRRR . . . 1
## See how VVPVGLRALVWVQR is shared by ECA3406 ECA3415 ECA3389 and
## links the three other componennts, namely ECA3399, ECA3389 and
## (ECA3415, ECA3406). Filtering that peptide out would split that
## protein group in three.
plotAdjacencyMatrix(ccomp[[3]])
#> Warning: vertex attribute color contains NAs. Replacing with default value 1
#> Warning: vertex attribute color contains NAs. Replacing with default value 1
#> Warning: vertex attribute color contains NAs. Replacing with default value 1
#> Warning: vertex attribute color contains NAs. Replacing with default value 1
#> Warning: vertex attribute color contains NAs. Replacing with default value 1
#> Warning: vertex attribute color contains NAs. Replacing with default value 1
#> Warning: vertex attribute color contains NAs. Replacing with default value 1
#> Warning: vertex attribute color contains NAs. Replacing with default value 1
#> Warning: vertex attribute color contains NAs. Replacing with default value 1
#> Warning: vertex attribute color contains NAs. Replacing with default value 1
#> Warning: vertex attribute color contains NAs. Replacing with default value 1
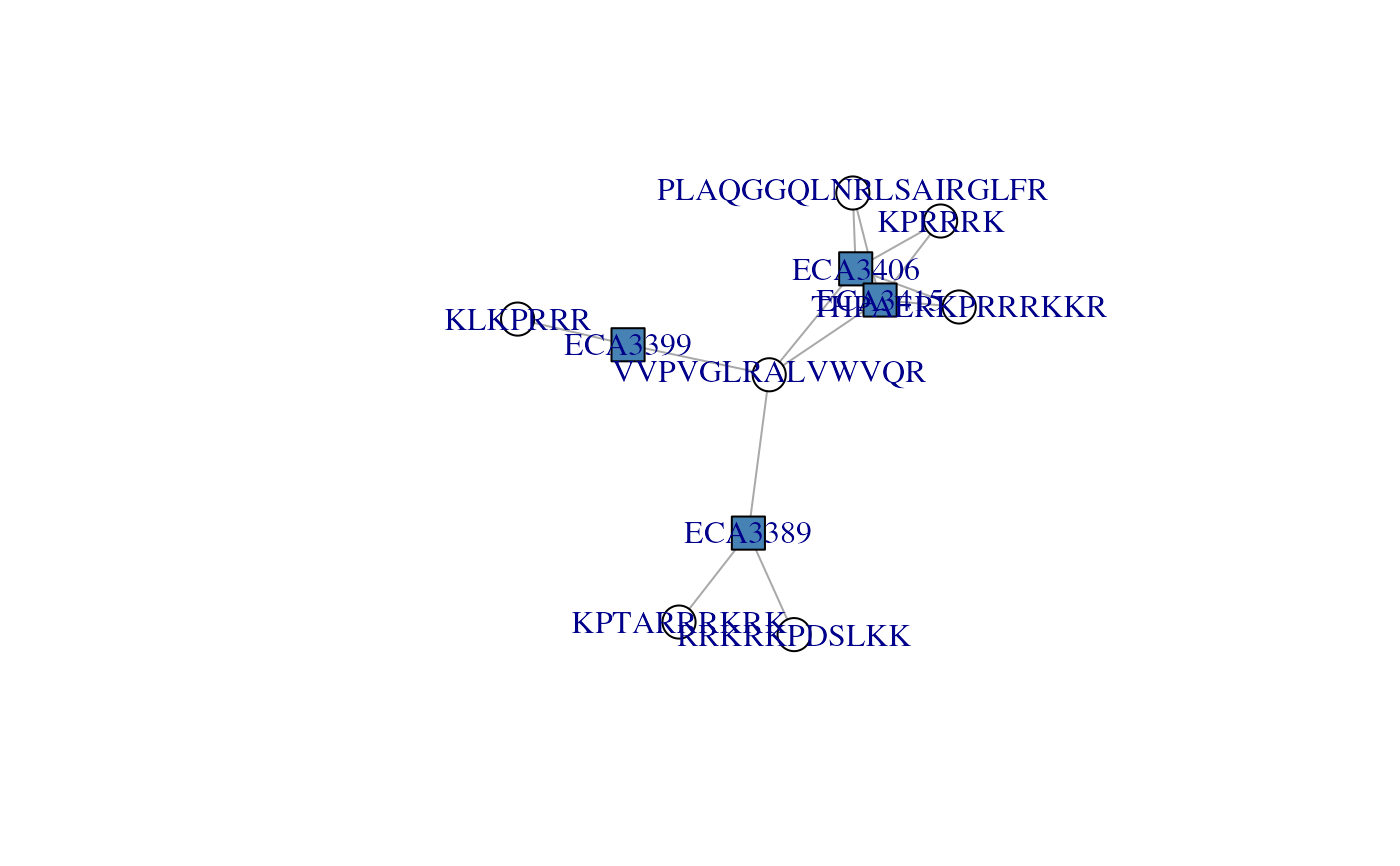 ## Colour protein node based on protein names similarity
plotAdjacencyMatrix(ccomp[[3]], 1)
## Colour protein node based on protein names similarity
plotAdjacencyMatrix(ccomp[[3]], 1)
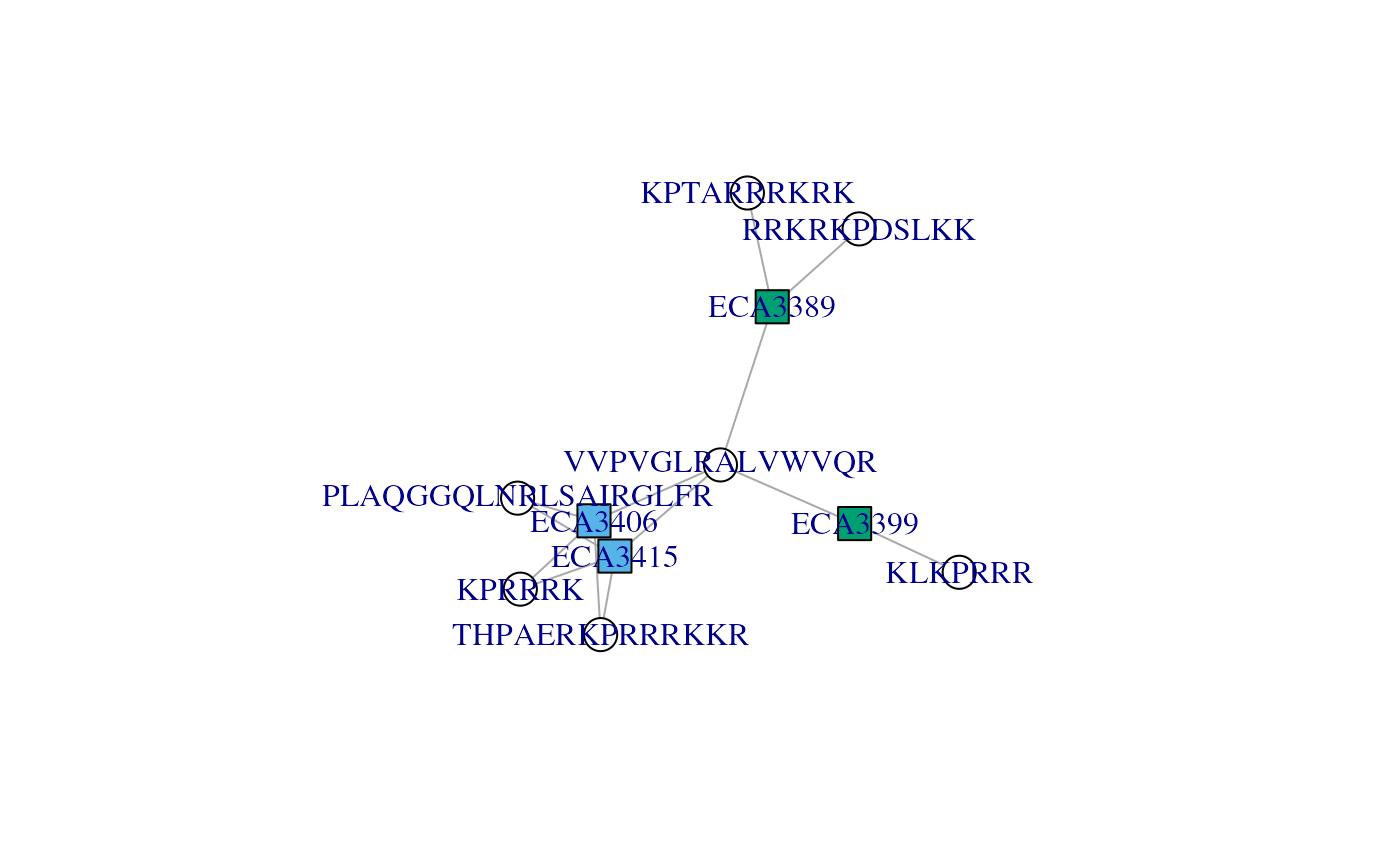 ## To select non-trivial components of size > 1
cc2 <- cc[ncols(cc) > 1]
cc2
#> An instance of class ConnectedComponents
#> Number of proteins: 49
#> Number of components: 21
#> Number of components [peptide x proteins]:
#> 0[1 x 1] 7[1 x n] 0[n x 1] 14[n x n]
## Use components features to prioritise their exploration
pri_cc <- prioritiseConnectedComponents(cc)
pri_cc
#> nrow ncol n_coms mod_coms n rs_min rs_max cs_min cs_max sparsity
#> 1082 7 4 3 0.4107143 14 1 4 2 4 0.5357143
#> 691 3 2 2 0.2187500 4 1 2 1 3 0.3333333
#> 1079 3 4 3 0.2142857 7 1 3 1 2 0.4166667
#> 920 4 2 2 0.1666667 6 1 2 3 3 0.2500000
#> 1078 2 2 2 0.1666667 3 1 2 1 2 0.2500000
#> 1080 2 2 2 0.1666667 3 1 2 1 2 0.2500000
#> 1359 2 2 2 0.1666667 3 1 2 1 2 0.2500000
#> 1423 2 2 2 0.1666667 3 1 2 1 2 0.2500000
#> 72 2 2 1 0.0000000 6 2 4 3 3 0.0000000
#> 526 2 2 2 0.0000000 4 2 2 2 2 0.0000000
#> 530 3 2 2 0.0000000 8 2 4 4 4 0.0000000
#> 1000 2 2 1 0.0000000 5 1 4 2 3 0.2500000
#> 1406 3 2 1 0.0000000 6 2 2 3 3 0.0000000
#> 1407 2 2 2 0.0000000 4 2 2 2 2 0.0000000
plotAdjacencyMatrix(connectedComponents(cc, 1082), 1)
## To select non-trivial components of size > 1
cc2 <- cc[ncols(cc) > 1]
cc2
#> An instance of class ConnectedComponents
#> Number of proteins: 49
#> Number of components: 21
#> Number of components [peptide x proteins]:
#> 0[1 x 1] 7[1 x n] 0[n x 1] 14[n x n]
## Use components features to prioritise their exploration
pri_cc <- prioritiseConnectedComponents(cc)
pri_cc
#> nrow ncol n_coms mod_coms n rs_min rs_max cs_min cs_max sparsity
#> 1082 7 4 3 0.4107143 14 1 4 2 4 0.5357143
#> 691 3 2 2 0.2187500 4 1 2 1 3 0.3333333
#> 1079 3 4 3 0.2142857 7 1 3 1 2 0.4166667
#> 920 4 2 2 0.1666667 6 1 2 3 3 0.2500000
#> 1078 2 2 2 0.1666667 3 1 2 1 2 0.2500000
#> 1080 2 2 2 0.1666667 3 1 2 1 2 0.2500000
#> 1359 2 2 2 0.1666667 3 1 2 1 2 0.2500000
#> 1423 2 2 2 0.1666667 3 1 2 1 2 0.2500000
#> 72 2 2 1 0.0000000 6 2 4 3 3 0.0000000
#> 526 2 2 2 0.0000000 4 2 2 2 2 0.0000000
#> 530 3 2 2 0.0000000 8 2 4 4 4 0.0000000
#> 1000 2 2 1 0.0000000 5 1 4 2 3 0.2500000
#> 1406 3 2 1 0.0000000 6 2 2 3 3 0.0000000
#> 1407 2 2 2 0.0000000 4 2 2 2 2 0.0000000
plotAdjacencyMatrix(connectedComponents(cc, 1082), 1)